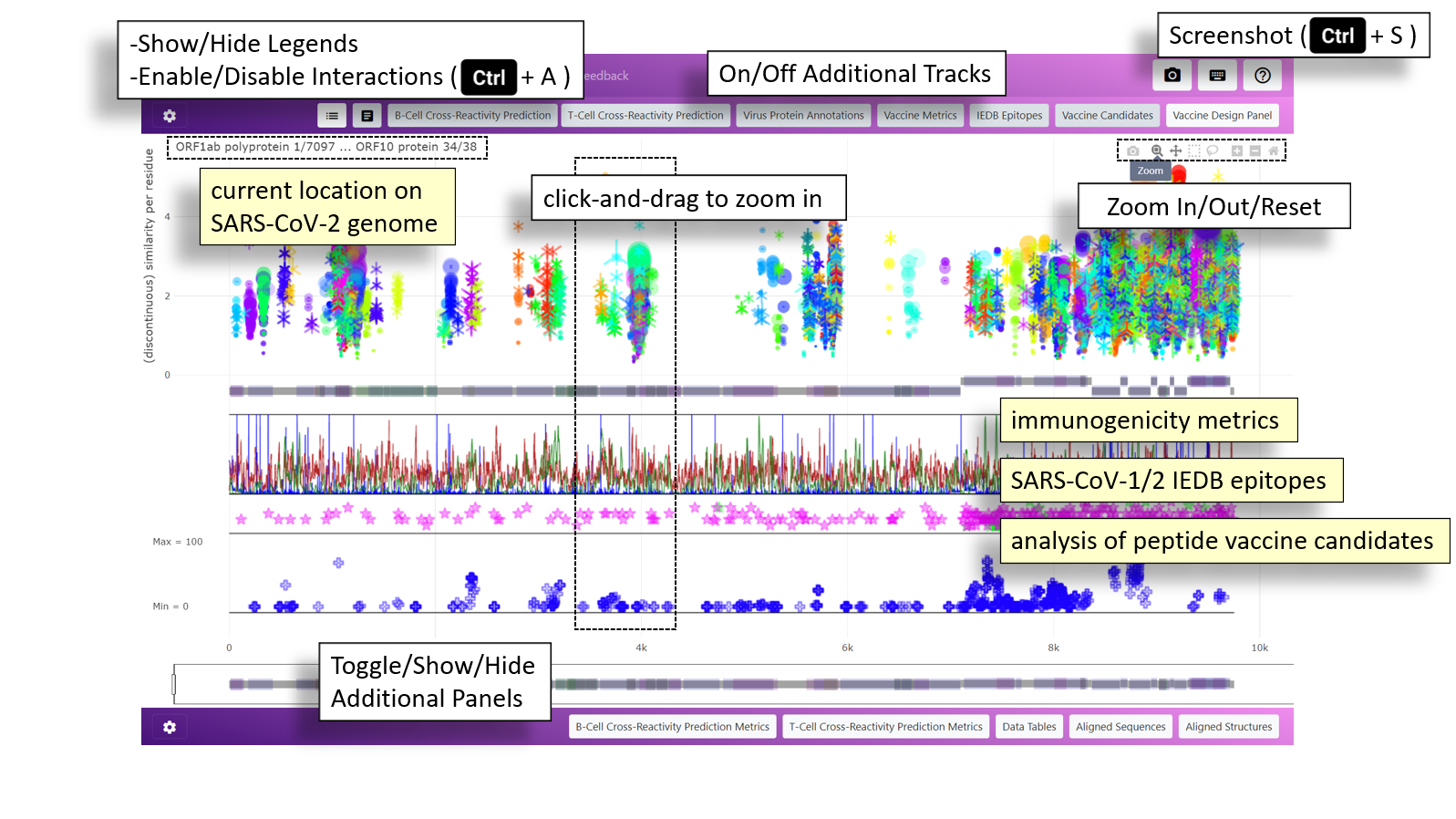
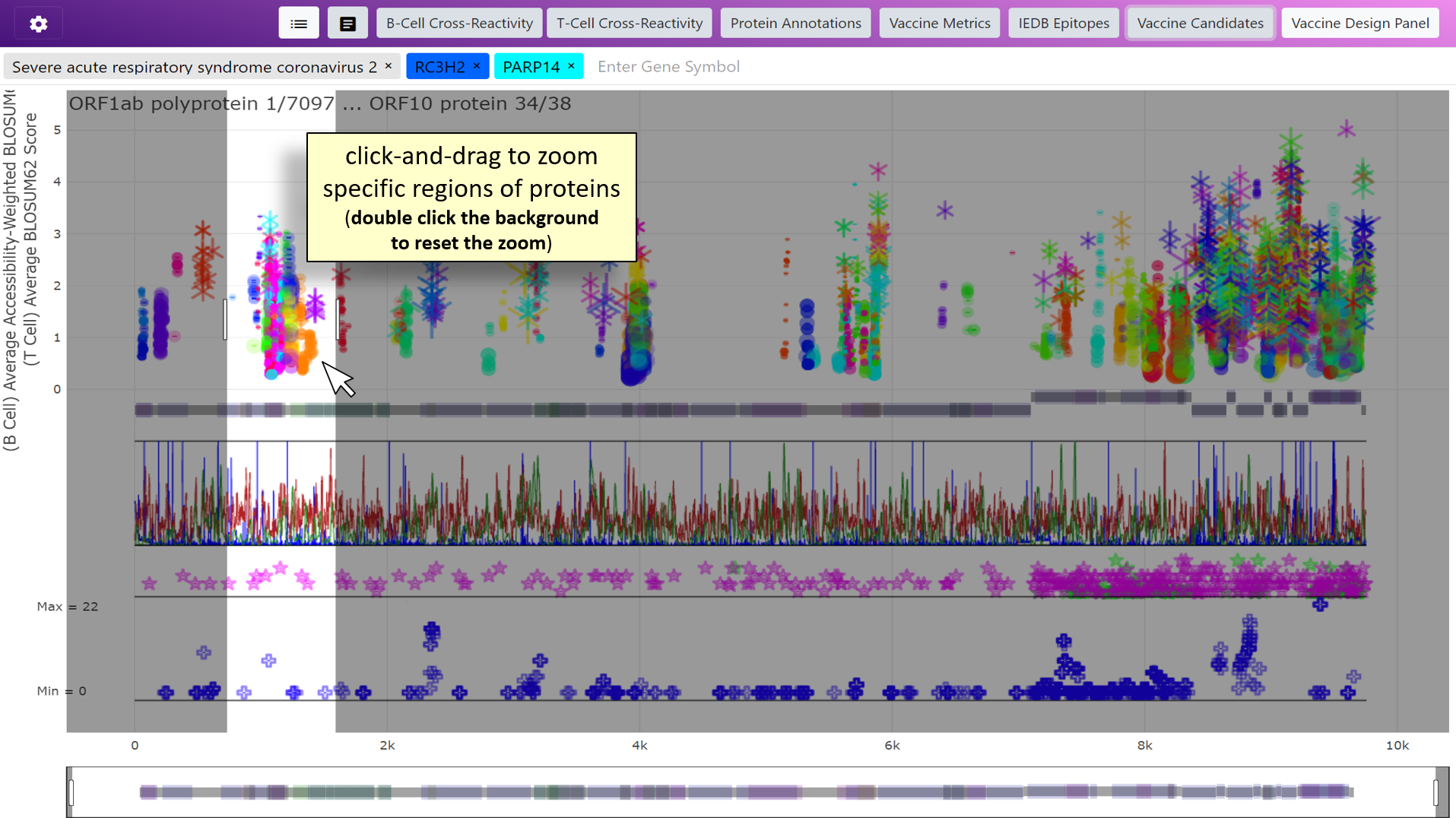
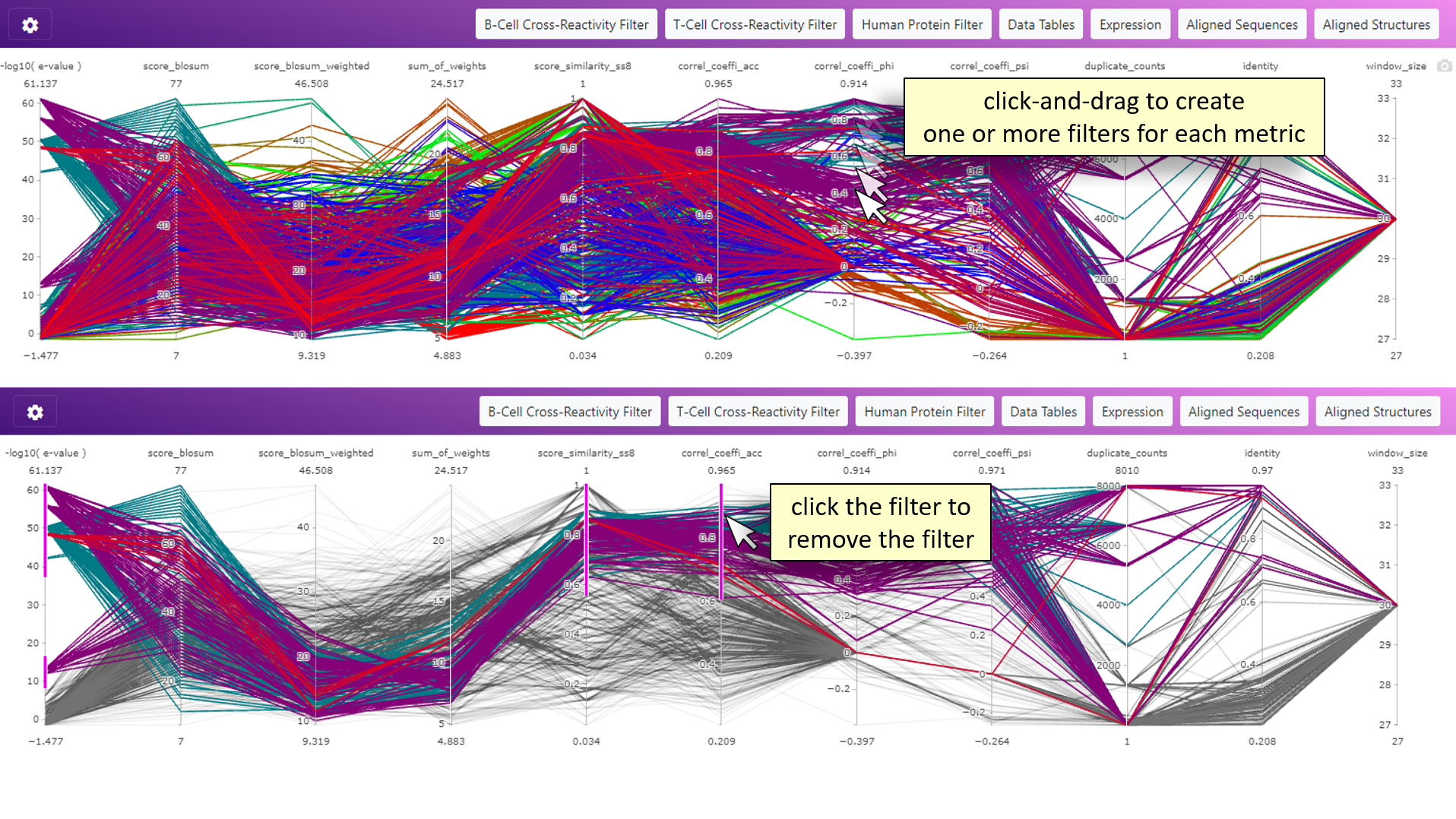
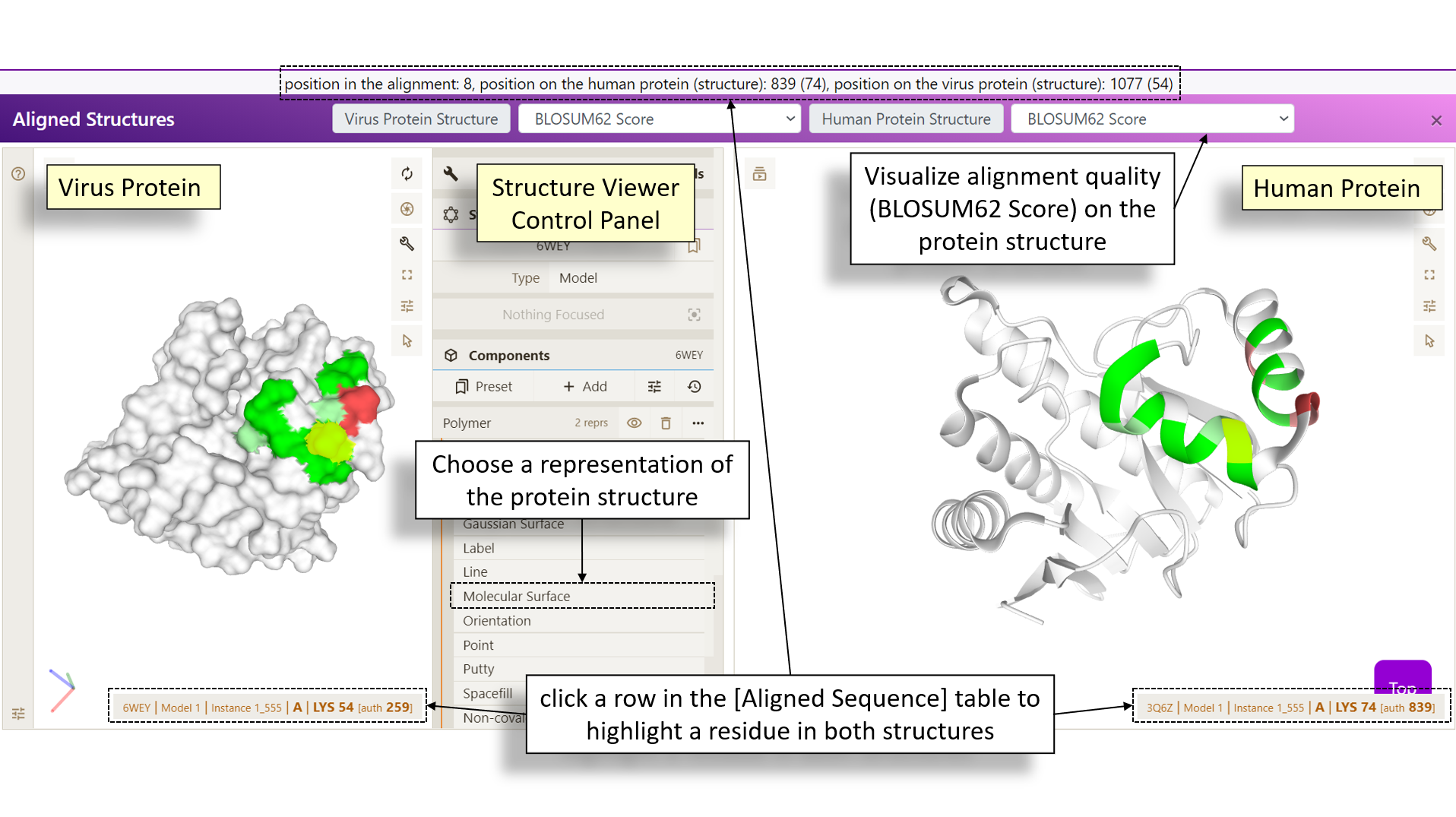
Introduction
Download Documentation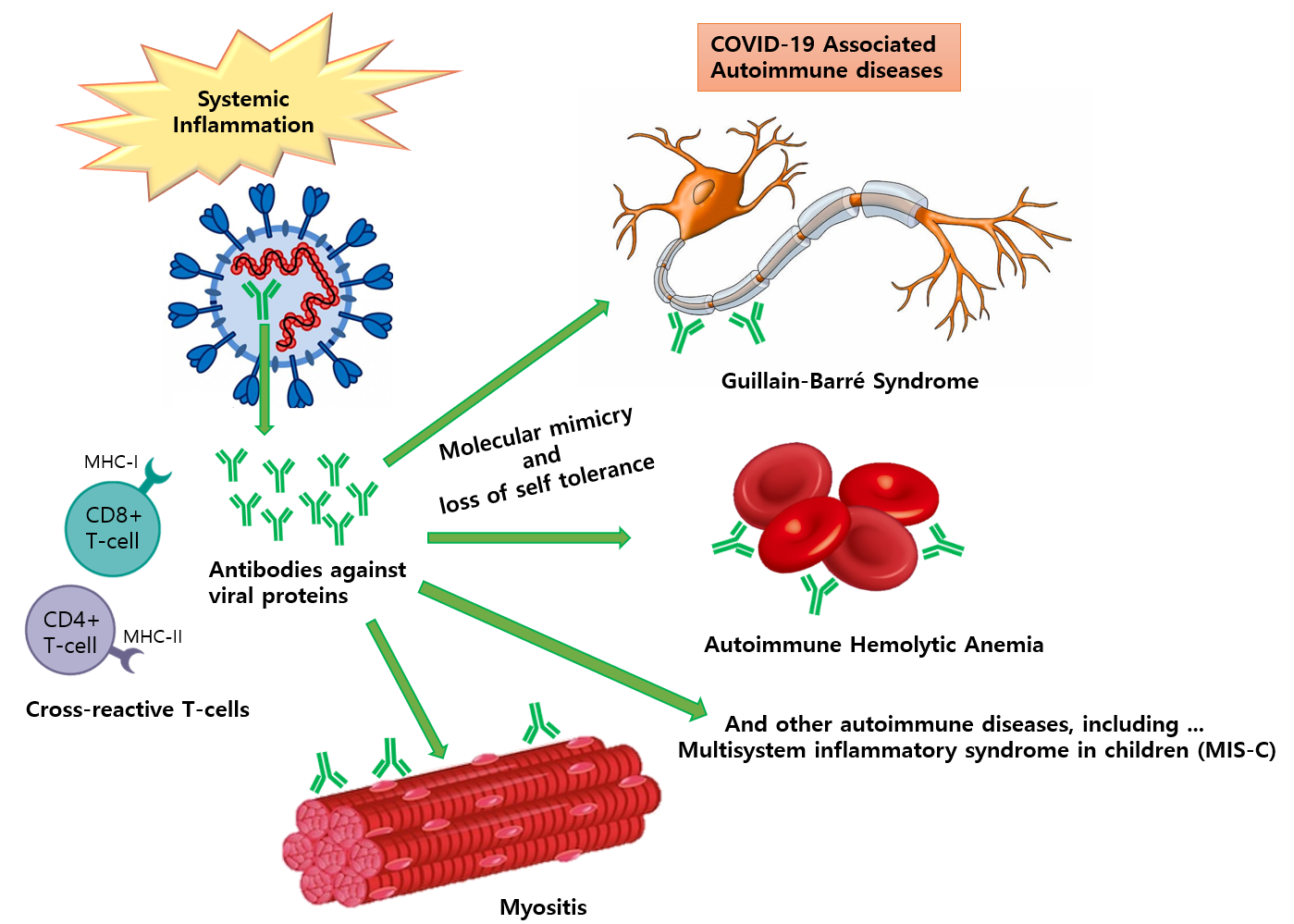
Abstract
The development of autoimmune diseases following SARS-CoV-2 infection, including multisystem inflammatory syndrome, has been reported, and several mechanisms have been suggested, including molecular mimicry. We developed a scalable, comparative immunoinformatics pipeline called cross-reactive-epitope-search-using-structural-properties-of-proteins (CRESSP) to identify cross-reactive epitopes between a collection of SARS-CoV-2 proteomes and the human proteome using the structural properties of the proteins. Overall, by searching 4 911 245 proteins from 196 352 SARS-CoV-2 genomes, we identified 133 and 648 human proteins harboring potential cross-reactive B-cell and CD8+ T-cell epitopes, respectively. To demonstrate the robustness of our pipeline, we predicted the cross-reactive epitopes of coronavirus spike proteins, which were recognized by known cross-neutralizing antibodies. Using single-cell expression data, we identified PARP14 as a potential target of intermolecular epitope spreading between the virus and human proteins. Finally, we developed a web application (https://ahs2202.github.io/3M/) to interactively visualize our results. We also made our pipeline available as an open-source CRESSP package (https://pypi.org/project/cressp/), which can analyze any two proteomes of interest to identify potentially cross-reactive epitopes between the proteomes. Overall, our immunoinformatic resources provide a foundation for the investigation of molecular mimicry in the pathogenesis of autoimmune and chronic inflammatory diseases following COVID-19.